Foire aux Questions
Logiciel Doric Neuroscience Studio
Pour que la version 6 reconnaisse les anciens appareils Doric, vous devez également mettre à jour le firmware de vos appareils. Instructions de téléchargement Ici. In cas où une mise à jour manuelle est conditions, vous pouvez retrouver le firmware spécifique à nos différents appareils sur cette page page web.
La Time Series fonctionnalité déplacée dans DNS v6. On le retrouve désormais sous le Paramètres globaux bouton dans le configuration languette (voir l'image ci-dessous).
Exporter dorique fichiers comme . Csv, vous devez utiliser le Éditeur de fichiers doriques module (voir image ci-dessous). En bref, en chargeant tous les fichiers de données doriques dans le module, vous pouvez cliquer sur « Exporter tout » pour les enregistrer sous forme de fichiers CSV.
Notes: si vous utilisez Matlab, Python ou Octave, vous pouvez lire directement .commec fichiers avec code à condition de ici. Avec une petite modification de votre pipeline d'analyse de données (quelques lignes de code au maximum), vous pouvez même remplacer le fichier .csv par .commec fichier. L'avantage de la .commec fichier (basé sur HBF5) is qu'il enregistre à la fois les données brutes et les paramètres d'enregistrement (utile pour le dépannage et/ou la reproductibilité). Nous ve déplacé vers ce format de fichier dans la version 6 car il peut gérer les métadonnées (vidéos de comportement, images, signal, TTL, etc.) et stocker très grande données de manière efficace.
danse™ et analyse de données
Vous en avez quelques différents Options pour analyser les données dans le .dorique Format:
-
Nous avons récemment sorti danse™ lequel est un logiciel conçu pour analyser.dorique filets. Plus précisément, avec PAS de codage conditions, danse™ peut:
-
Traitement de base (supprimer les artefacts, décimer, interpolation linéaire, calculer dF/F0, trouver des pointes, etc.)
-
Importez des événements de stimuli/comportements et des vidéos de comportement à partir d'autres appareils
-
Calculez les événements comportementaux liés à l'activité neuronale (y compris le suivi des animaux, calculez la présence des animaux dans les zones, la distance des animaux par rapport aux points, le score de mouvement et créez des événements comportementaux à partir de toutes ces mesures comportementales à l'aide d'un seuil réglable)
-
Créer des tracés qui combinent des données sur l'activité neuronale et le comportement (par exemple, histogrammes temporels péri-événement/péri-stimuli)
-
Automatisere pipelines de traitement et d’analyse de données sans codage conditions
Noter la Onglet Support technique de notre site Web maintenant contient de nombreuses tutoriel danse™ vidéos qui peut vous donner une idée de ce que le logiciel peut faire. Actuellement, seul le contenu lié à la photométrie a été créé sous forme de didacticiel, mais nous ajoutons régulièrement à cette vidéothèque des vidéos de didacticiel sur le comportement et la microscopie.s à venir. Si cela vous intéresse option, contactez-nous au sales@doricenses.com pour un essai gratuit de 15 jours et/ou un devis. Nous proposons également des démonstrations virtuelles via zoom, y compris une visite guidée utilisant vos propres données avec nous pour voir comment vous pouvez utiliser au mieux le logiciel pour analyser vos expériences spécifiques.
-
Logiciel Doric Neuroscience Studio (DNS), notre logiciel gratuit d'acquisition de données, contient le Analyseur de signal, Analyseur d'image, ainsi que Analyseur de comportement des modules qui incluent des outils de traitement de données de base également proposés par danse™ (comme calculer dF/F, trouver des pointes, etc.).
-
Si vous souhaitez utiliser Matlab, Python, R ou Octave, vous pouvez directement lire .dorique fichiers avec le code fourni sur nos GitHub dépôt. Ceci comprend la .dorique sortie de la Analyseur de signal, Analyseur d'image, ainsi que Analyseur de comportement modules. Cela vous permet de faire des analyses plus approfondies sur Photométrie dF/ F0 calculs pour créer vos tracés et calculer vos statistiques dans l'un ou l'autre ceux ,softwares.
Photométrie à fibre
Un guide utile présentant et comparant nos différents systèmes de photométrie est disponible ici. Ce guide, en plus de passer en revue leurs spécificités, pourra également vous guider vers un système qui pourrait mieux correspondre à votre propre expérience. Si vous avez besoin d'aide supplémentaire, contactez l'un de nos spécialistes au sales@doricenses.com.
La Onglet Support technique de notre site Web maintenant contient plusieurs vidéos tutorielles fournir de l'aide avec les systèmes de photométrie installation et mise en place. Si vous avez besoin de plus assistance avec sconfiguration de votre système, contactez un de nos spécialistes au sales@doricenses.com.
Comparaison entre entrelacé et Verrouillage modes :
|
Mode entrelacé |
Mode de verrouillage |
|
|
Fréquence temporelle maximale |
60 Hz |
1000 Hz |
|
Systèmes de photométrie compatibles |
Base et forfait |
De base uniquement |
|
Nombre de signaux |
2 (de base) jusqu'à 3 (lot) |
Jusqu'à 4 |
|
Sensible à la lumière ambiante |
OUI |
NON |
La Imode entrelacé autres deux excitations LEDs, comme présenté dans l’image ci-dessous.
La Mode de verrouillage utilise des fréquences de référence sinusoïdales pour piloter 2 et plus encore Excitations LED à différentes fréquences (Voir l'image ci-dessous). alors a algorithme de démodulation séparés les signaux.
Cette méthode a plusieurs avantages:
- Les signaux démodulés sont invariants à la lumière ambiante et au bruit au-dessus/en dessous de la fréquence de référence
- Have pas d'artefact marche/arrêt puisque les LED oscillent toujours entre Vmin et Vmax (et ne sont jamais complètement éteints). Le seul inconvénient du mode verrouillage est qu'il nécessite une fréquence d'échantillonnage élevée, ce qui n'est pas toujours possible, notamment lorsque l'on utilise des capteurs CMOS au lieu de photodétecteurs pour enregistrer les signaux photométriques.
Pas de signal: (des creux et des bossesre presque identique entre trace expérimentale et trace isobestique)
Signal faible (des bosses dans le signal fonctionnel do ne se produit pas dans la trace isobestique mais de faible amplitude)
Signal fort (les bosses dans le signal fonctionnel ne se produisent pas dans le tracé isobestique)
Selon le système, le niveau d'expression des fluorophores enregistrés, et un tas de facteurs externes, ces valeurs seront toujours différentes et doivent être optimisé pour vos conditions spécifiques. Un bon point de départ est de rechercher des articles sur la région cérébrale d’intérêt et/ou qui ont utilisé votre fluorophore. La puissance des LED est généralement indiquée et peut vous donner une idée d'une plage de puissance plus spécifique pour votre boîtier. Nous recommandons bien que que la puissance isobestique soit la moitié de la puissance du signal expérimental. Cela réduira le photoblanchiment. Vous devez mesurer la puissance LED de chaque LED une à la fois (éteindre la LEDs dans le module Driver LED).
Il y a deux possibilités: soit les paramètres ne sont pas optimaux ou il y a un dysfonctionnement composant dans le système.
-
Pour FMC ou iFMC systèmes (avec LED externes)
-
Vérifiez les connexions du système. Assurez-vous notamment le Les clés du connecteur sont bien aligné dans le emplacements pour prises, surtout lors du serrage du écrou d'accouplement (voir image ci-dessous). Un mauvais alignement peut réduire considérablement la transmission à travers la fibre et entraîner de fortes baisses de puissance.
-
Réglez le pilote LED en mode CW et en mode faible consommation avec un courant maximum (200 mA). to reproduire les puissances minimales et maximales indiquées sur la LED et iFMC Fiche de données. S'ils diffèrent de plus de 10%, contactez le support Doric (support@doriclenses.com) pour garantir le bon fonctionnement de l'appareil.
-
ilFMC Systèmes Gen.2 (avec LED intégrées, mais pilote de LED séparé)
-
Tournez la bague de réglage (bague en argent) dans le sens des aiguilles d'une montre pour augmenter la puissance transmise via l'atténuateur variable (Voir l'image ci-dessous)
-
Vérifiez les connexions des systèmes. Assurez-vous notamment le Les clés du connecteur sont bien aligné dans le emplacements pour prises, surtout lors du serrage du l'écrou d'accouplement (voir image ci-dessous). Un mauvais alignement peut réduire considérablement la transmission à travers la fibre et entraîner de fortes baisses de puissance.
-
Utilisez un wattmètre et un 400µm Cordon de brassage NA0.57 avec le courant réglé à 200 mA pour reproduire le minimum et le maximum les chiffres de puissance indiqués sur le ilFMC Fiche de données. S'ils diffèrent davantage supérieur à 10%, contactez le support Doric (support@doriclenses.com) pour assurer la dispositif travaille correctement.
-
Systèmes ilFMC Gen.3 (avec LED intégrées et pilote de LED)
-
Vérifier les connexions des systèmes
-
Utilisez un wattmètre et un 400µm Cordon de brassage NA0.57 avec le courant réglé à 500 mA pour reproduire la puissance maximale indiquée sur le ilFMC Fiche de données. S'ils diffèrent de plus de 10%, contactez le support Doric (support@doriclenses.com) pour assurer la ilFMC fonctionne correctement.
Étant donné qu'environ 80 % du signal photométrique provient du volume de tissu situé at une profondeur de ~ 200 um de la facette de la fibre, la valeur approximative zone de champ de vision pour chaque diamètre de noyau est directement relié au diamètre du cordon de brassage. (Vous pouvez simplement calculer la surface de chaque cordon de brassage en utilisant le diamètre pour voir la différence de champ de vision.) Le NA du cordon de brassage modifiera la forme et la profondeur des collections de données. Vous pouvez en apprendre davantage sur l'effet du diamètre du noyau et de l'AN sur la collecte de données photométriques dans ce document. papier. Notez que les gens utilisent des cordons de brassage de diamètres différents en fonction de la taille de leur région d'intérêt. Si votre région d'intérêt a a diamètre d'environ 200 um, utilisez le cordon de brassage 200 um. Si c'est plus grand, utilisez le 400µm core à la place. Vous obtiendrez 75 % de signal en plus en utilisant a 400µm puisque vous pouvez imager beaucoup plus de neurones avec ce cordon de brassage de plus grande surface. Pour la photométrie à fibre une NA plus grande est recommandée pour collecter plus de signal ; Alors que iIl est vrai qu'une NA plus faible augmentera la profondeur de pénétration, lorsque nous recommandons une fibre à NA élevée, nous supposons que la canule est placée à proximité (moins de quelques µm) à la zone de détection. Ensuite, puisque la position peut être manuellement optimisé, nous recommandons l'optimisation la collection de fluorescence avec une fibre à haute NA.
-
L'une des causes des artefacts de mouvement sur le signal est la transmission optique de la jointure tournantet qui peut varier en rotation.
-
Si le rat génère une accélération rapide, il peut être difficile de corriger ces artefacts de mouvement.
-
Le système embarqué sur le rotor d'un joint tournant électrique assisté par couple, Que l' RFMC au sein de l’ RBFMC, supprimera ces variations de signal, car le capteur est directement sur le rotor et ne passe donc pas par un joint rotatif optique.
-
D'autres causes d'artefact de mouvement sont les pertes par courbure des fibres et le mouvement de la fibre. implant. La perte de courbure des fibres est difficile à éviter. Vous pouvez envisager d'utiliser une gaine plus résistante, c'est-à-dire une gaine ARMO au lieu de la gaine de 1.1 mm. hytrel veste pour le cordons de brassage à l'animal. Il est moins flexible et affectera davantage le comportement, mais il est acceptable pour les rats. Et cela aidera le câble à survivre plus longtemps.
-
Le mouvement de l'implant fibreux nécessite une bonne cimentation et peut-être une protection autour de l’implant pour garantir sa stabilité. Pour les souris, c'est moins of un problème, mais pour les rats, nous savons que les chercheurs ajoutent certaines fonctionnalités pour solidifier l'implant. Vous pouvez envisager en utilisant la virole de 2.5 mm au lieu de 1.25 mm, ou un autre type de prise. Vous pouvez jetez un oeil ha dit que pour des informations plus détaillées en ce qui concerne les implants que nous proposons.
Minicubes avec LED intégrées réduire le nombre de connexions optiques et l'encombrement global du système, ce qui simplifie globalement le système. Elle offre cependant moins de flexibilité à l’utilisateur en termes de sources lumineuses à utiliser. D'autre part, nsur-intégré minicubes représentent plus grand nombre de composants et d'espace, mais la modularité augmente la flexibilité Afin que tu puisses utiliser différentes LED / lasers et/ou fluorophores dans le futur que par réplacingurgiter quelques composants conditions au lieu d'acheter un tout nouveau système.
REMARQUE: il n'y a pas prix différence jusqu'à XNUMX fois an mini-cube intégré et mini cube non intégré avec LED et câbles acheté séparément.
Alors que le point isosbestique des fluorophores GCaMP a été bien caractérisé et que les 405 et 415 nm sont largement utilisés en photométrie verte, la photométrie rouge en est encore à ses balbutiements. Certains n'utilisent pas du tout de point isobestique pour l'une ou l'autre couleur de photométrie (même si c'est le cas). une belle façon pour contrôler la fluorescence de fond/les artefacts, en particulier chez les animaux en mouvement libre).
WNous recommandons généralement d'utiliser le Mode CC. Quand à Mode CC, le signal de la photodiode est directement transmis à l'amplificateur sans altération. La plage de sortie au niveau du connecteur BNC est de 0 V à 5 V. Ce mode est recommandé pour les expériences de photométrie et doit être utilisé par défaut, sauf indication contraire. En mode AC, le signal de la photodiode passe à travers un filtre passe-haut de 30 Hz avant d'être envoyé à l'amplificateur. Toutes variations inférieures à 30 oscillations par seconde ainsi que la valeur moyenne du signal sont supprimées. La plage de sortie au niveau du connecteur BNC est de -5 V à 5 V. Ce mode offre une plage dynamique deux fois supérieure mais peut masquer un état de saturation du préamplificateur et doit être utilisé avec précaution.
Le commutateur de gain sélectionne entre trois ou deux (selon les générations de mini-cubes) niveau de sortie pour la même lumière de fluorescence d’entrée. Le gain 1x transmet le signal du préamplificateur directement au BNC tandis que les 10x et 100x ajoutent 2 autres niveaux d'amplification. Gagner sélection ne doit être effectué que pour un réglage très grossier du signal. Le gain doit être diminué si l'amplificateur est saturé (à la ligne autour de 5 V) et augmenté s'il est inférieur à 0.5 V. Il est assez courant que le Amplification 10X est utilisé pour les expériences de photométrie, mais cela dépendra également de votre signal.
-
FRJ_1x1_PT : 2 % de crête à crête
-
FRJ_2x2_PT : 3 % de crête à crête
-
AFRJ_2x2_PT : 3 % de crête à crête
Microscopie
Pour microscopes en utilisant une connexion USB-3 à l'ordinateur (eTOSM, eTSM-Gén3):
-
Veillez à ce que Entraînement du microscoper est branché sur l'ordinateur à l'aide du connecteur fourni. Câble USB-3.
-
Assurez-vous que le Connecteur de câble électrique est branché sur le appareil approprié (ordinateur et microscope).
-
Lorsque le pilote du microscope 1 couleur est activé, le OnL'interrupteur /Off doit être allumé ON et la LED blanche clignote lors de l'initialisation. Si la lumière reste allumée sans clignoter lors de la première mise sous tension, redémarrez le pilote du microscope.
Pour les microscopes en utilisant une connexion Ethernet à l'ordinateur (gestion durable des forêts, 2CFM):
-
Assurez-vous que le pilote du microscope est branché à l'ordinateur à l'aide d'un câble Ethernet.
-
Assurez-vous que le connecteur du câble électrique est branché sur le appareil approprié. Le pilote du microscope doit être relié au microscope.
-
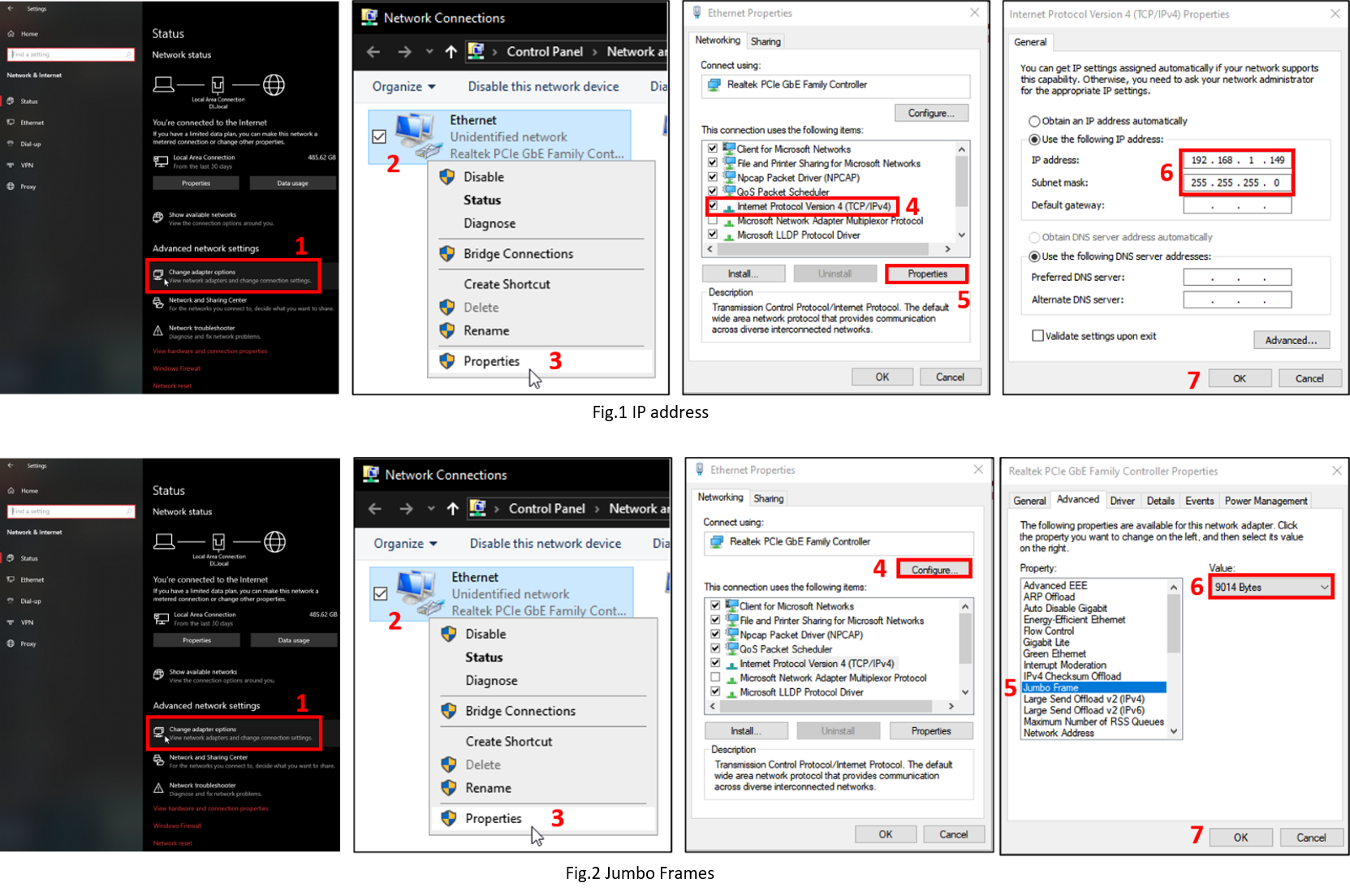
Assurez-vous que l'adresse IP est statique (voir Fig.1)
-
Assurez-vous que les trames Jumbo sont activées (voir Fig.2)
-
Le pare-feu Windows peut empêcher la communication. Pour vous assurer que la communication n'est pas bloquée, ouvrez le Fenêtre de configuration du Pare-feu Windows, puis cliquez sur Autoriser une application via le pare-feu. De là, sélectionnez le Bouton Modifier les paramètres, trouver Neurosciences doriques Studio ,software et vérifier la Privé et public cases à cocher.
-
Dans le Centre de réseau et de partage, vérifiez la connexion Ethernet ; cela devrait indiquer Réseau non identifié. Si Câble réseau débranché s'affiche malgré le branchement du câble Ethernet et la mise sous tension du pilote, désactiver et réactivez la connexion Ethernet.
-
Assurez-vous que le réseau et le partage sont correctement configurés à 1 Gbit/s en double-cliquant sur la connexion Ethernet et vérifier la vitesse.
-
Lorsque le pilote du microscope 1 couleur est activé, le OnL'interrupteur /Off doit clignoter en bleu lors de l'initialisation. Si la lumière est maintenu sans aucun clignotement lors de la première mise sous tension, redémarrez le pilote du microscope.
Si vous avez besoin de plus assistance avec cela, contactez un de nos spécialistes au support@doriclenses.com.
Le pilote du microscope doit être connecté à un port Ethernet de l'ordinateur. Si un adaptateur USB vers Ethernet est utilisé pour une autre fonction, telle que l'accès à Internet, l'adaptateur doit être désactivé lors de la première initialisation du microscope.
Lors de l'exécution d'une expérience de microscopie avec Doric Neuroscience Studio, nous vous recommandons deactivatingurgiter tout internet-en utilisant des programmes pouvant entrer en conflit avec Doric Neuroscience Studio (c'est à dire Skype, pare-feu, etc)
De plus, nous vous recommandonssingurgiter un ordinateur avec these Caractéristiques:
-
Système d'exploitation : Windows 10 ou 11
-
Processeur : Quad Core I7 3.46 GHz
-
RAM: 16 Gb
-
Carte graphique dédiée : avec Open GL version 4.6 recommandée
-
Ordinateur de bureau recommandé
Enfin, WLes fenêtres peuvent limiter les performances pour réduire la consommation d'énergie.
Pour s'assurer que la communication n'est pas limité, ouvrez l'option Alimentation fenêtre et suivez ces étapes:
-
Appuyez sur les touches Windows + R pour ouvrir la boîte de dialogue Exécuter.
-
Tapez le texte suivant : « powercfg.cpl", Puis appuyez sur Entrée.
-
Dans la fenêtre Options d'alimentation, sous Sélectionner un mode d'alimentation, choisissez Hautes performances.
-
Si vous ne voyez pas le Optimisation option, cliquez sur la flèche vers le bas à côté de Afficher supplémentaire plans.
-
Si disponible, modifiez le Système en attente ainsi que Le système hiberne paramètres à Jamais.
-
Cliquez Enregistrer les modifications ou cliquez sur OK.
*Les images perdues sont des images noires qui se produisent lorsqu'une image est perdue lors de la communication. Ils peuvent facilement être repérés dans Intensité moyenne in Suivi du retour sur investissement si la valeur descend à 0.
Les images obtenu avec Logiciel Doric Neuroscience Studio sont enregistrés dans a .dorique format, qui est un format de type HDF5. Ils peuvent être visualisés à l'aide ,softwares prenant en charge le format HDF5 tel que Logiciel Doric Neuroscience Studio (utilisant l' Analyse d'imagesr plugin), ImageJ (Fonction d'importation et divers plugins) ou une visionneuse HDF. Code exemples sont également à condition de ici sur notre site Internet ainsi que permettre à lire les images en Python, matlab et Octave.
Nous recommandons que le Cordon de brassage à fibre optique est de longueur égale ou inférieure à celle du Câble électrique pour microscope quand je me connecteed à la Joint tournant opto-électrique assisté. Même si le câble est bouclé, la distance entre le joint tournant et Le connecteur du cordon de brassage doit être plus court que la longueur du câble électrique.
Ces deux composants sont destinés à être collés ensemble après l'installation. S'ils n'ont pas été collés pendant Lors de l'installation, ajoutez une goutte de colle à séchage rapide sur la bordure entre la canule et l'anneau de réglage de la saillie.
Remplissez l'intérieur de la canule avec KWIK-CAST (WPI) pour faire office de capuchon. Après avoir retiré le mastic séché, nettoyez la surface extérieure de la Rod Lens à l'aide d'un coton-tige légèrement imbibé d'alcool isopropylique.
- Assurez-vous que les pinces du microscope sont suffisamment desserré (visser le canon).
- Assurez-vous que la canule et le corps du microscope sont correctement alignés.
- Vérifiez l'installation de la canule Des instructions . Quelques conseils sont disponibles dans ce note d'application. Vous pouvez également jeter un oeil à ceci vidéo d'instructions sur le microscope unicolore or this vidéo pour microscope bicolore.
-
Nous vous recommandons de vous assurer d'abord que le corps du microscope est correctement connecté à la plaque de base de l'implant, car un petit écart peutL'un et l'autre peuvent affecter la transmission du signal et/ou l'éclairage.
-
Habituellement, le système peut distinguer les cellules 2 ou 3 semaines après l'implantation de la canule. Néanmoins, une meilleure qualité d’image est obtenue 2 à 8 semaines après l’implantation. Le temps d'attente pour la réparation des tissus peut être une bonne période pour utiliser le microscope factice pour entraîner l'animal à supporter l'encombrement d'un microscope sur sa tête et à s'habituer à se déplacer facilement dans sa cage avec lui. Si vous réalisez 2 à 3 images après l'implantation, envisagez d'attendre encore 10 jours-2 semaines avant de réessayer. If le problème persiste, pensez à vérifier le positionnement de l'implant dans l'échantillon par rapport à l'emplacement des cellules fluorescentes à imager (pour vérifier s'ils sont hors de portée de la distance de travail du microscope)
Dans la dernière version de Doric Neuroscience Studio v5, un nouveau format de fichier, .dorique format, a été introduit, et ce format est le seul format à être enregistré dans les dernières versions v6. Les données sont exclusivement enregistrées en HDF5 fichiers .doric format d'extension (les formats .csv et .tiff à partir de la version 5 ne sont pas est plus disponible). Les fichiers HDF 5 peuvent être lus avec Matlab, Octave et Python. Des exemples de code sont fournis pour faciliter analyse des données avec ces applications externes.
L'utilisation de .doric format de fichier par rapport aux autres formats fournit plusieurs avantages :
-
Stocke des types de données hétérogènes (tels que les vecteurs de signaux, des images, et vidéos) dans un seul fichier;
-
Enregistre les paramètres de configuration utilisés pour enregistrer les données
-
Pas de longueur maximale, ce qui est idéal pour les enregistrements de séries chronologiques durer plus d'une journée et/ou des enregistrements avec vidéos comportementales
-
Intégrez en toute transparence comportement données dans vivoneuralr enregistrements
-
Compatible avec le nouveau logiciel d'analyse de données de Doric, danseTM
-
Facilement lire .doric à Matlab, Python ou Octave utilisant à condition de codes.
Une remarque importante est que les configurations de canaux enregistrées à partir de la version 5 ne sont plus compatibles avec la version 6.
Optogénétique
Pour l'optogénétique, cela dépend du cas d'utilisation : (1) voulez-vous que la lumière atteigne une zone aussi grande que possible ? (ie pour une manipulation importante et donc le plus grand changement de comportement affectant) ou (2) la précision de la lumière dans une petite zone est requise (pour éviter de stimuler les zones voisines afin d'éviter toute confusion). Alors que a une NA inférieure augmentera la profondeur de pénétration.
Le choix de la fibre optique NA sera également une influence utilisée pour effectuer la stimulation lumineuse. Lors de l'utilisation d'une source de lumière à diode laser, des cordons de brassage NA 0.22 sont couramment utilisés, le NA du laser étant environ de 0.15.
Caméras comportementales
Les caméras N&B de Doric sont compatibles avec les régions de lumière visible et proche infrarouge (pas de filtre IR). SVoir la sensibilité spectrale ci-dessous. Vous pouvez vérifier ha dit que pour des informations plus détaillées sur la caméra comportementale que nous proposons.
Joints rotatifs
Vérifiez que l'alimentation câblee du joint tournant n'est PAS branché sur le port USB d'un ordinateur, mais sur le fourni source de courant. Le port USB ne fonctionne pasmettre fin au minimum conditions courant à alimenter adéquatement les moteurs du joint tournant.
Si cela ne résout pas le problème, contactez Support dorique (support@doriclenses.com).
-
Vérifiez les connexions du système. Assurez-vous notamment le Les clés du connecteur sont bien aligné dans le emplacements pour prises, surtout lors du serrage du l'écrou d'accouplement (voir image ci-dessous). Un mauvais alignement peut réduire considérablement la transmission à travers la fibre et entraîner une ruptureLa puissance chute.
-
Réglez le pilote LED en mode CW ainsi que définir la courant max - 500A. Mesurez la puissance maximale du joint rotatif seul et avec un patch cordon, et le comparer à puissance de sortie de le cordon de brassage seul. Si l' les mesures différer by plus de 10% du signaler la transmission sur le feuille de test, contactez le support Doric (support@doriclenses.com) pour garantir le bon fonctionnement de l'appareil.
